4. PSF Extraction#
From the theory lecture, you learned PSF (point spread function) is the shape of the point source in your detector, after passing through all the optics. It includes the diffraction pattern, seeing (natural + telescope), etc. For space telescopes, PSF will be very stable because both the seeing sources will be negligible (if thermal expansion is well suppressed). Only the gradual change due to the degradation of the optics maybe important (or some dramatic changes by dust impact).
In reality, it is practically impossible to know the PSF a priori for ground-based observations, especially because both the seeing factors are chaotic and impossible to predict. Also, there is absolutely no guarantee that the PSF will be the same for the consecutive images (you may need to include a slowly varying time-variation term, such as that in Stetson 1987) Thus, one must rely on empirical estimation of the PSF of the given image.
Students living in the 2020s may want to ask the AI to estimate the PSF in the image. However, as in any other branches of science, the current AI technology is not suitable for most of the quantitative analyses (especially because of the error analysis). Thus, classical approach would be
Tell the program a rough FWHM value (“initial guess”).
Tell the program which shape of PSF you expect (e.g., “use Gaussian fitting”, “use Moffat and spline interpolation”, etc.).
Manually check if the PSF is well estimated (check the “residuals”).
Manually check if the PSF photometry is done correctly (repeat the processes if necessary).
4.1. Preparation#
First, load the data:
from pathlib import Path
import _tool_visualization as vis
import numpy as np
import pandas as pd
import ysfitsutilpy as yfu
import ysphotutilpy as ypu
from astropy.io import fits
from astropy.nddata import CCDData, Cutout2D
from astropy.stats import sigma_clipped_stats
from astropy.table import Table
from matplotlib import pyplot as plt
from matplotlib import rcParams
from photutils.aperture import CircularAnnulus, CircularAperture
from scipy.interpolate import RectBivariateSpline
plt.style.use('default')
rcParams.update({'font.size':12})
DATAPATH = Path("../../Tutorial_Data/")
LOGPATH = Path("./tmp") # a temporary output directory
LOGPATH.mkdir(exist_ok=True, parents=True)
PSF_CENTROIDING_THRESH_SIGMA = 3.
PSF_CENTROIDING_AREA_FRAC = 1/4
PSF_CENTROIDING_SKY_ANNUL_FACTORS = (4., 6.)
PSFSIZE_FACTOR = 4
# Load our data
fpath = DATAPATH/"SNUO_STX16803-M11-1-1-20190507-171622-V-60.0.fits"
ccd_raw = CCDData.read(fpath)
gain = ccd_raw.header.get("GAIN", 1)
rdnoise = ccd_raw.header.get("RDNOISE", 0)
Status messages could not be retrieved
WARNING: FITSFixedWarning: 'datfix' made the change 'Set MJD-OBS to 58610.719699 from DATE-OBS'. [astropy.wcs.wcs]
PSF_CENTROIDING_THRESH_SIGMA(k): The threshold level for PSF stars’ centroiding process. The pixels above the k-σ error-bar (variance = pixel variance + sky variance) will be used in SExtractor-like algorithm.PSF_CENTROIDING_AREA_FRAC: The minimum fraction of the cutout area used for centroiding process (\((PSFSIZE+1)^2\)), that is required for the PSF star to occupy.PSF_CENTROIDING_SKY_ANNUL_FACTORS: The inner/outer radii of the sky estimating annulus for PSF stars, in the unit of determined FWHM.
Then, do the cosmic-ray rejection. (see CR rejection lecture)
NOTE:
yfu.crrejis nothing butastroscrappy.detect_cosmics, with some additional tasks to add history in the FITS files.
crpath = LOGPATH / "SNUO_STX16803-M11-1-1-20190507-171622-V-60.0_CR.fits"
if crpath.exists():
ccd = CCDData.read(crpath)
else:
ccd, crmask = yfu.crrej(
ccd=ccd_raw,
gain=gain,
rdnoise=rdnoise,
objlim=5.,
sepmed=True, # IRAF LACosmic is sepmed=False
)
ccd.write(crpath)
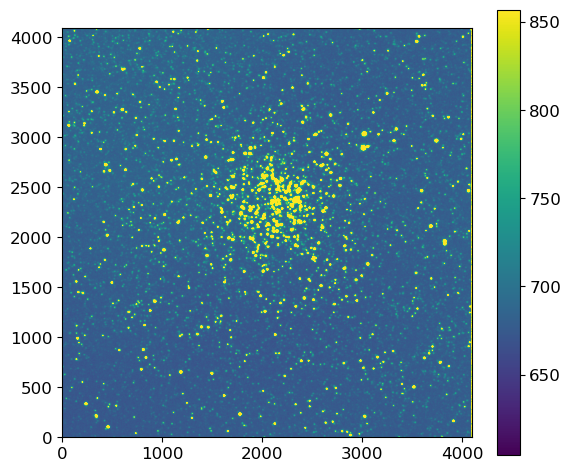
fig, ax = plt.subplots(1, 1, figsize=(6, 5))
im = vis.norm_imshow(ax, ccd.data, zscale=True)
plt.colorbar(im)
plt.tight_layout()
4.2. Initial FWHM#
From ginga, you have to find the FWHM of the stars.
Open ginga
Operation→FBrowser→ open your FITS file (Here,LOGPATH/SNUO_STX16803-M11-1-1-20190507-171622-V-60.0_CR.fits)Operation→Analysis→PickFind a star that (1) does not overlap with others, (2) is bright enough but not saturated. They are not at the center of the image.
left click and right click to do “pick” (See official documentation of Pick)
Bottom right panel:
Settings→ setRadiusto 30 pix (since SNUO 1m has too small pixel scale…) &&FWHM fittingtoMoffatClick
Redo PickTop right panel: Select
Radial.Check if it is good:
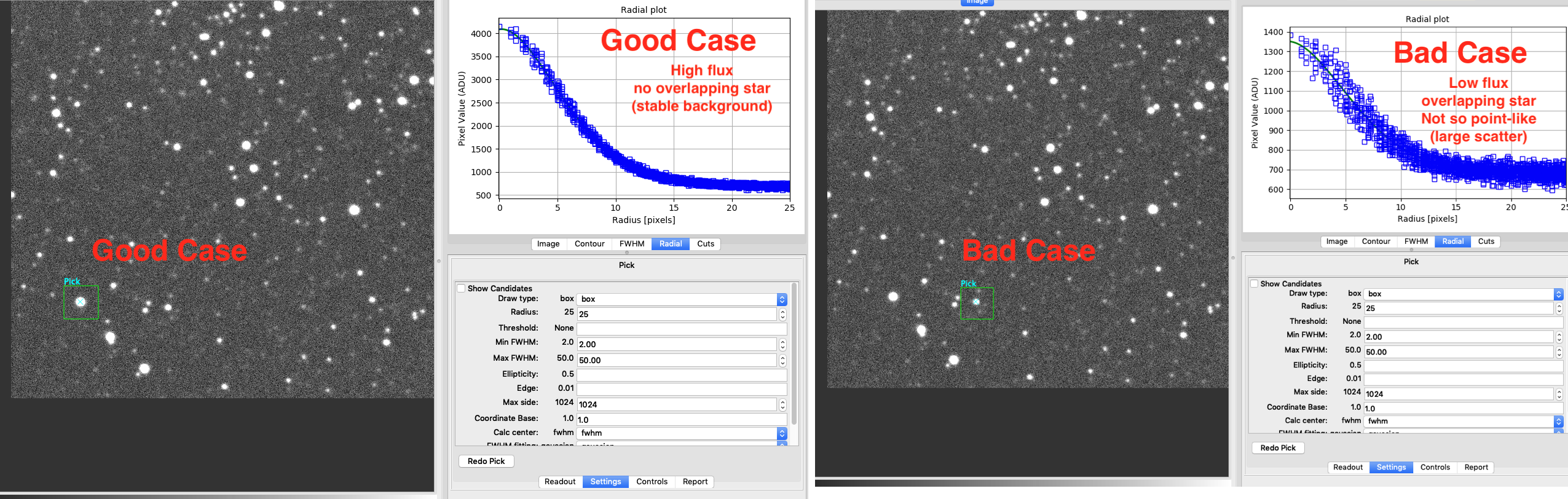
If not good, select a different star.
Bottom right panel:
Report→Add PickDo this for around 10 stars throughout the image (better if regularly spaced).
Save tablewith namepick_log.csv.
4.3. Load the Pick Results#
Load the ginga Pick results, and find the centers of the stars.
_pick_log_path = LOGPATH/"pick_log.csv"
# get column names
with open(_pick_log_path, "r") as f:
pick_colnames = f.readline().strip().split(" ")[1:] # First one is just `#`
pick_log = pd.read_csv(_pick_log_path, names=pick_colnames, sep=" ", comment="#")
# NOTE: ginga pick has 1-indexing: https://github.com/ejeschke/ginga/issues/769
pick_log["x"] -= 1
pick_log["y"] -= 1
# Take sigma-clipped median of FWHM as initial guess
FWHM_INIT = sigma_clipped_stats(pick_log["fwhm"], sigma=3.0, maxiters=5)[1]
PSFSIZE = PSFSIZE_FACTOR*FWHM_INIT + 1
print(f"FWHM_INIT = {FWHM_INIT:.2f} pixels")
print(f"PSFSIZE = {PSFSIZE} pixels")
pick_log[["x", "y", "fwhm", "brightness", "background"]]
FWHM_INIT = 12.03 pixels
PSFSIZE = 49.13712434103719 pixels
| x | y | fwhm | brightness | background | |
|---|---|---|---|---|---|
| 0 | 3863.020138 | 707.683315 | 11.962709 | 756.294478 | 703.163513 |
| 1 | 3514.125040 | 500.461007 | 12.047357 | 3113.724378 | 723.208466 |
| 2 | 791.043571 | 676.635925 | 12.041618 | 1806.826784 | 703.645050 |
| 3 | 238.905269 | 332.319899 | 12.026944 | 3362.454716 | 721.233795 |
| 4 | 880.530746 | 1262.908716 | 11.953833 | 1600.975989 | 708.042664 |
| 5 | 450.723343 | 2016.108091 | 11.974557 | 2423.947034 | 715.987640 |
| 6 | 802.402071 | 2468.025588 | 11.724034 | 2934.561519 | 698.899902 |
| 7 | 776.520519 | 3882.919494 | 11.921826 | 1851.317307 | 699.666199 |
| 8 | 1600.434322 | 3353.373197 | 11.943629 | 2051.583265 | 697.423889 |
| 9 | 3738.857578 | 2961.212126 | 12.383431 | 8053.690733 | 703.893646 |
| 10 | 3591.739585 | 2460.098066 | 12.371278 | 3526.151165 | 698.613129 |
| 11 | 3682.802596 | 2106.829978 | 12.307477 | 6108.703287 | 701.417389 |
| 12 | 3514.123997 | 500.461282 | 12.089511 | 3149.151973 | 693.987732 |
| 13 | 1496.579152 | 637.077055 | 12.137889 | 2415.422264 | 689.493286 |
Here,
fwhmis the FWHM from radial profile (you must have chosen radially symmetric objects, not galaxy-like objects)backgroundis the local background level (rough estimate)brightnessis the “peak pixel value” - “background”.FWHM_INITis the sigma-clipped median of the FWHM values of the selected point-sources.
From now on, I will set the PSF size to be a box of side length PSFSIZE = 4*FWHM_INIT+1 ~ 49. This means, when estimating PSF, I extract 49x49 pixels around the center of each star. Therefore, ideally, there should be no overlapping stars or background object in the nearby ~ 25 pixels (~ 49/2).
4.4. Prapare Cutouts for Each Star#
Now cut the 49x49 pixels around the stars. Because the central location from ginga pick is not the most accurate, I will
Re-calculate the central pixel using sep.
# Initialize
pick_fit = dict(x0=[], y0=[], x=[], y=[], shift=[], fwhm=[], bkg_ginga=[])
# Calculate sky values for each star
_ans = CircularAnnulus(
np.array([pick_log["x"], pick_log["y"]]).T,
r_in=FWHM_INIT*PSF_CENTROIDING_SKY_ANNUL_FACTORS[0],
r_out=FWHM_INIT*PSF_CENTROIDING_SKY_ANNUL_FACTORS[1]
)
_skys = ypu.sky_fit(ccd, _ans)
for c in _skys.colnames:
pick_fit[c] = np.array(_skys[c])
# prepare errormap
var = yfu.errormap(ccd, gain_epadu=gain, rdnoise_electron=rdnoise, return_variance=True)
pick_cuts = []
for i, row in pick_log.iterrows():
fwhm = row["fwhm"]
x0 = row["x"]
y0 = row["y"]
msky = pick_fit["msky"][i]
ssky = pick_fit["ssky"][i]
# cutout PSFSIZE+1 pixels (in case bezel is not enough for centroiding)
_cut = Cutout2D(ccd.data, (x0, y0), PSFSIZE + 1)
_varcut = Cutout2D(var.data, (x0, y0), PSFSIZE + 1)
# SExtractor-like extraction
obj, seg = ypu.sep_extract(
_cut.data,
thresh=PSF_CENTROIDING_THRESH_SIGMA, # k-sigma detection
bkg=msky, # background level
var=_varcut.data + ssky**2, # variance map including sky
minarea=PSF_CENTROIDING_AREA_FRAC*PSFSIZE**2, # minimum area
)
pos = _cut.to_original_position((obj["x"].iloc[0], obj["y"].iloc[0]))
# Add results to pick_fit
shift = np.sqrt((pos[0] - x0)**2 + (pos[1] - y0)**2)
for k, v in zip(["x0", "y0", "x", "y", "shift", "fwhm", "bkg_ginga"],
[x0, y0, pos[0], pos[1], shift, row["fwhm"], row["background"]]):
pick_fit[k].append(v)
pick_cuts.append(Cutout2D(ccd.data, pos, PSFSIZE))
pick_fit = pd.DataFrame.from_dict(pick_fit)
# Aperture photometry for stars for future reference
aps, ans = ypu.circ_ap_an(
np.array([pick_fit["x"], pick_fit["y"]]).T,
r_ap=PSFSIZE/2,
r_in=FWHM_INIT*PSF_CENTROIDING_SKY_ANNUL_FACTORS[0],
r_out=FWHM_INIT*PSF_CENTROIDING_SKY_ANNUL_FACTORS[1]
)
pick_phot = ypu.apphot_annulus(ccd, aps, ans, gain=gain, rdnoise=rdnoise, error=np.sqrt(var))
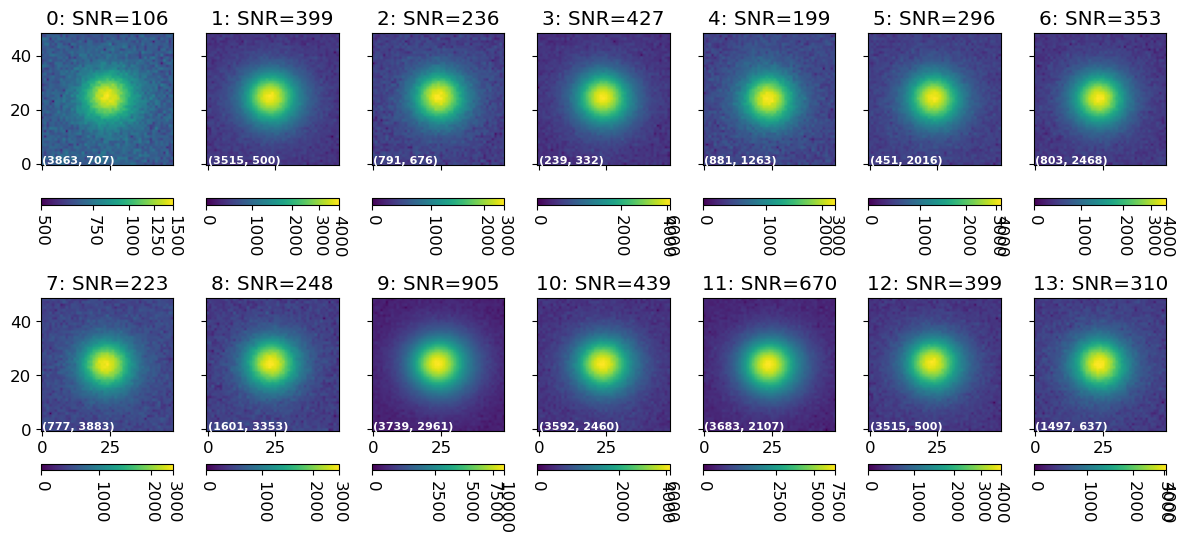
ncol = 7
nrow = max(len(pick_log) // ncol, 1)
fig, axs = plt.subplots(nrow, ncol, figsize=(12, 2.8 * nrow), sharex=True, sharey=True)
for i, _cut in enumerate(pick_cuts):
irow = i // ncol
_ax = axs[irow, i - irow * ncol]
_row_phot = pick_phot.iloc[i]
_ax.set_title(f"{i}: SNR={_row_phot['snr']:.0f}")
im = vis.norm_imshow(_ax, _cut.data, stretch="sqrt")
cb = vis.colorbaring(fig, _ax, im)
pos_str = "({:.0f}, {:.0f})".format(pick_fit.iloc[i]['x'], pick_fit.iloc[i]['y'])
_ax.text(0, 0, pos_str, fontsize=8, fontweight='bold', color='w')
plt.setp(cb.ax.get_xticklabels(), rotation=-90)
plt.tight_layout()
plt.show()
4.5. Construct PSF Model#
pick = Table.read(, format="ascii.csv",
delimiter=' ', data_start=2, comment="#", names=pick_colnames)
# NOTE: ginga pick has 1-indexing: https://github.com/ejeschke/ginga/issues/769
pick["x"] -= 1
pick["y"] -= 1
# Only pixels near the stars in the txt file
mask = np.ones_like(data).astype(bool)
for row in pick:
pos = (row["x"], row["y"])
iy, ix = Cutout2D(mask, position=pos, size=PSFSIZE).bbox_original
mask[iy[0]:iy[1], ix[0]:ix[1]] = False
thresh = np.median(data)
print("Finding stars...")
finder = DAOStarFinder(fwhm=FWHM_INIT, threshold=thresh, exclude_border=True)
found = finder(data, mask=mask)
print(f"Stars initially selected for the PSF estimation: {len(pick)}")
if len(pick) != len(found):
print("!!!!!! pick and found length not match !!!!!!")
found.remove_columns(["npix", "sky"]) # remove confusing columns from DAOStarFinder
Cell In[6], line 1
pick = Table.read(, format="ascii.csv",
^
SyntaxError: invalid syntax